一些常见的R语言命令,主要是基于Baser,后面我会继续整理基于R语言dplyr包处理数据的技巧方法
0.目录
- 1.Load R packages
- 2.set path & import data
- 3.Identify missing values
- 4.conditional assignment
- 5.tapply (grouped calculation)
- 6.quantile and summary
- 7.origin-data (coefplot in stata)
- 8.dataclear in hs8-year level (average®)
- 9.dataclear in hs8-year level
- 10.scatter
- 11.density
- 12.summarize the data
- 13.regress
- 14.reg R versus stata
- 15.dataclear for bar & line
- 16.bar
- 17.line
Table of contents generated with markdown-toc
1.Load R packages
library(ggplot2)
library(tidyr)
library(RColorBrewer)
library(dplyr)
library(ggstatsplot)
2.set path & import data
rm(list=ls())
setwd("D:/project/Feb2023_Lumpy/data")
dir()
load("project_data3.RData")
head(data)
data <- as.data.frame(data)
data$lnP <- log(1+data$P)
data$lnQ <- log(1+data$Q)
data$lnV <- log(1+data$V)

3.Identify missing values
data$lnpif<-is.na(data$lnP)
data$lnqif<-is.na(data$lnQ)
data$lnvif<-is.na(data$lnV)
data$ifNA <- complete.cases(data)
data_heal <- data[data$lnqif==FALSE&data$lnvif==FALSE&data$lnpif==FALSE, c("id","hs8","Year","capt","T_hs08") ]
4.conditional assignment
data_heal$type[data_heal$capt == 1 & data_heal$Year >= 2007 ] <- "hg2"
data_heal$type[data_heal$capt == 1 & data_heal$Year < 2007 ] <- "hg3"
data_heal$type[data_heal$capt != 1 & data_heal$Year >= 2007 ] <- "hg1"
data_heal$type[data_heal$capt != 1 & data_heal$Year < 2007 ] <- "hg4"
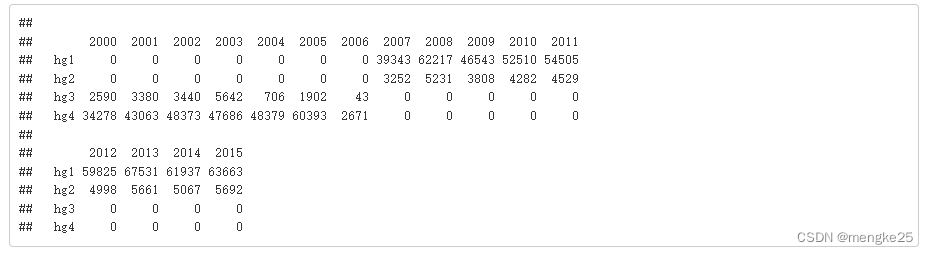
table(data_heal$type,data_heal$Year)
5.tapply (grouped calculation)
data_heal_ <- data[is.na(data_heal$T_hs08) == FALSE & is.nan(data_heal$T_hs08) == FALSE , c("id","hs8","Year","capt","T_hs08") ]
data_heal_$group_check <- as.double(runif(dim(data_heal_)[1]))
data_heal_$group[data_heal_$group_check <= 0.25] <- "1"
data_heal_$group[data_heal_$group_check > 0.25 & data_heal_$group_check <= 0.5] <- "2"
data_heal_$group[data_heal_$group_check >0.5& data_heal_$group_check <= 0.75] <- "3"
data_heal_$group[data_heal_$group_check >0.75] <- "4"
data_heal_$lnT <- log(1+ data_heal_$T_hs08)
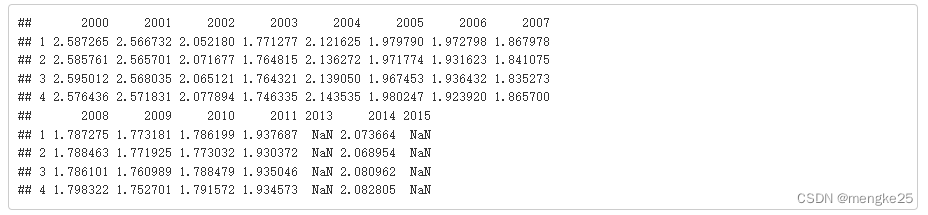
tapply(data_heal_$lnT,data_heal_$group,"mean",na.rm = TRUE)
mean(data_heal_$lnT[data_heal_$group == "2"],na.rm = TRUE)
tapply(data_heal_$lnT,list(data_heal_$group,data_heal_$Year),"mean",na.rm = TRUE)
# manager <- c(1, 2, 3, 4, 5)
# country <- c("US", "US", "UK", "UK", "UK")
# gender <- c("M", "F", "F", "M", "F")
# age <- c(32, 45, 25, 39, 99)
# leadership <- data.frame(manager, country, gender, age)
# tapply(leadership$age, leadership$country, mean) # 求在不同country水平下的age的均值


6.quantile and summary
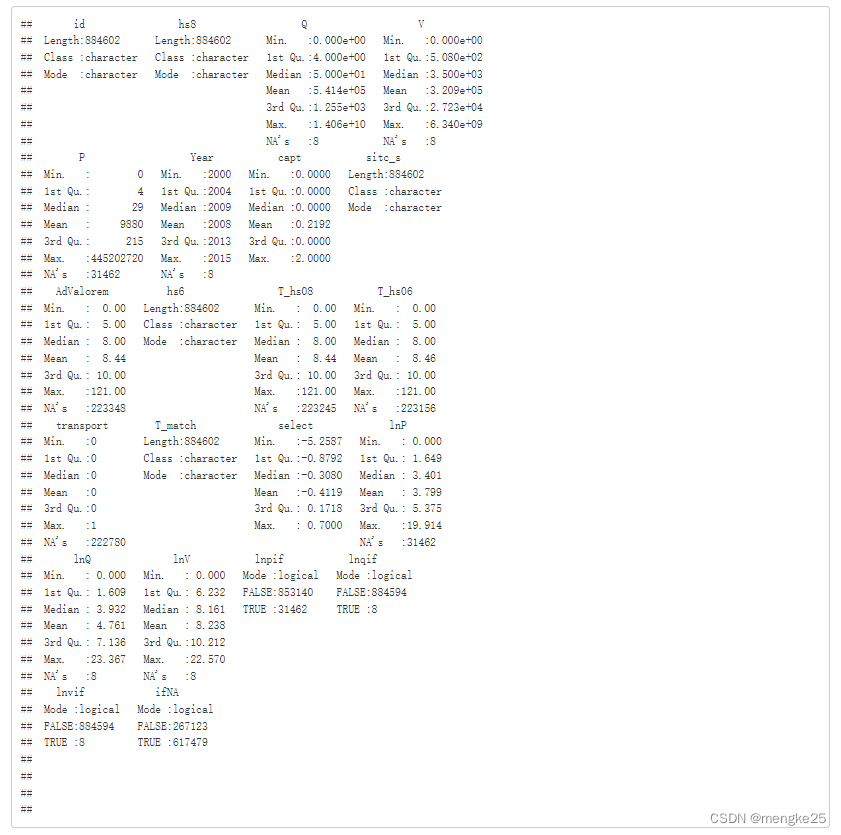
summary(data,maxsum = 20, quantile.type = 7)
quantile(data$lnP,seq(from = 0 ,to = 1 , by = 0.1),na.rm = TRUE)
7.origin-data (coefplot in stata)
fitmod_T <- estimatr::lm_robust(T_hs08 ~ as.factor(Year) , data = data_heal[data_heal$capt !=0, ], se_type= "stata")
summary(fitmod_T)
ggcoefstats(fitmod_T,exclude.intercept = TRUE,stats.labels = FALSE)
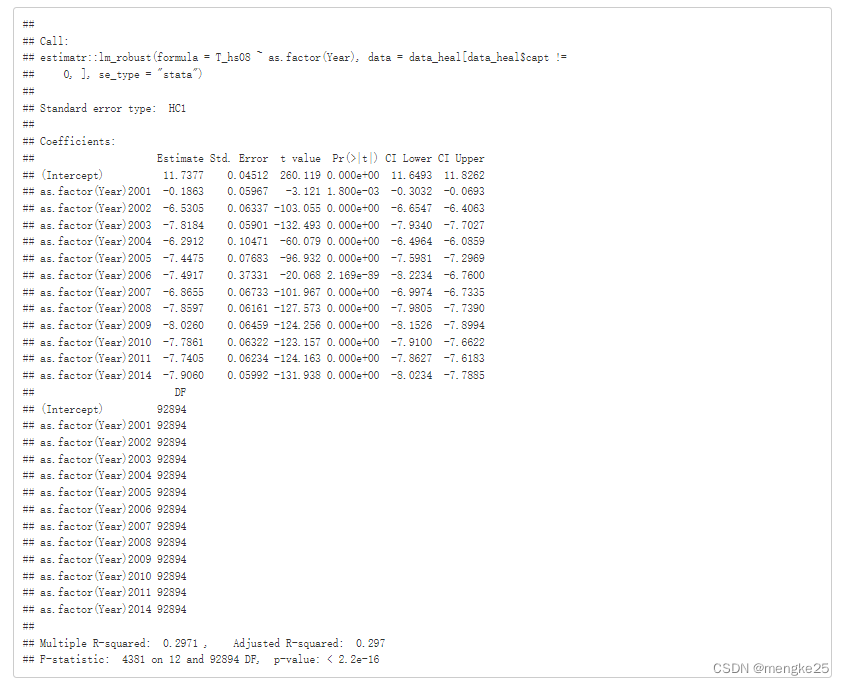
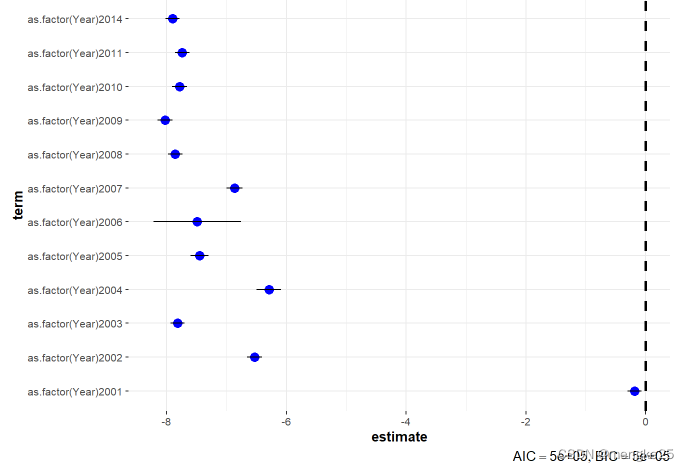
8.dataclear in hs8-year level (average®)
# average for T_hs08
data_heal2 <- aggregate(data_heal$T_hs08,list(data_heal$hs8,data_heal$Year),mean)
# rename
names(data_heal2) <- c("hs8","year","meanT")
# Extract non-missing values
data_heal2 <- data_heal2[is.na(data_heal2$meanT) != TRUE, c("hs8","year","meanT")]
# Logarithmic
data_heal2$logT <- log(1+data_heal2$meanT)
# regress
fitmod_T2 <- estimatr::lm_robust(logT ~ as.factor(year),data = data_heal2, se_type = "stata")
summary(fitmod_T2)
# plot the coef
ggcoefstats(fitmod_T2,exclude.intercept = TRUE,stats.labels = FALSE,vline = TRUE)
boxplot(data_heal2$logT ~ as.factor(data_heal2$year))
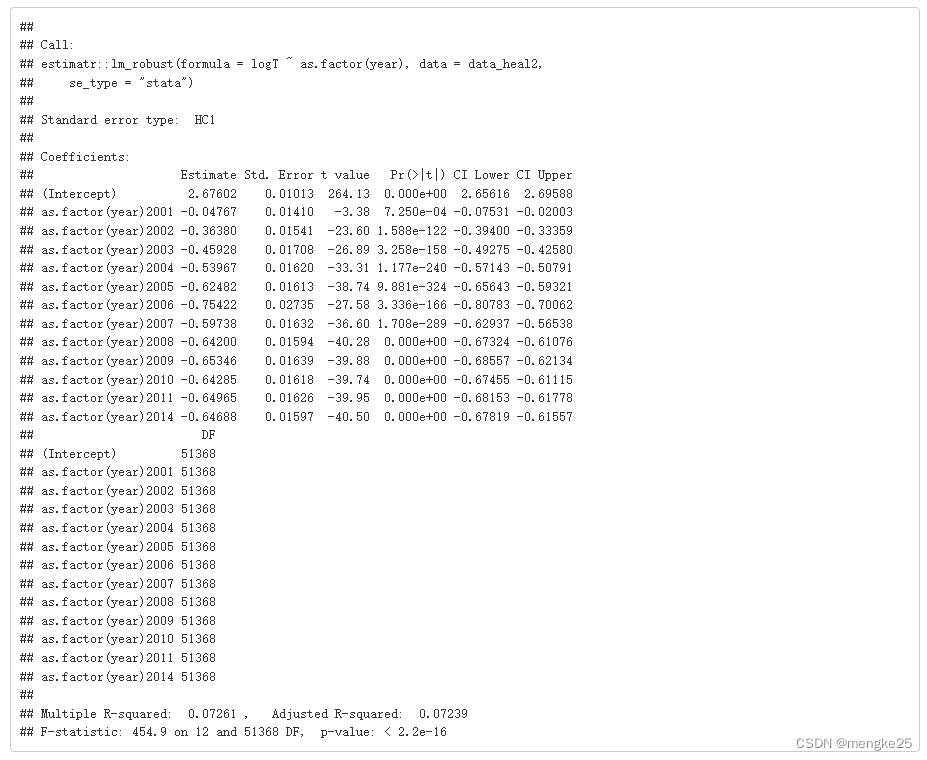
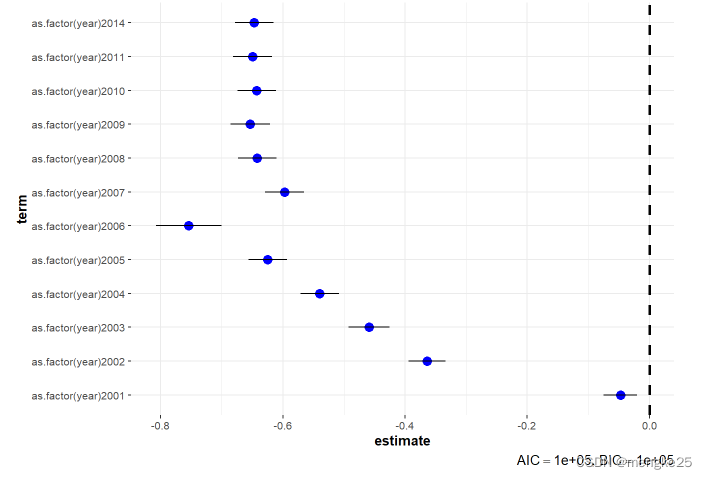
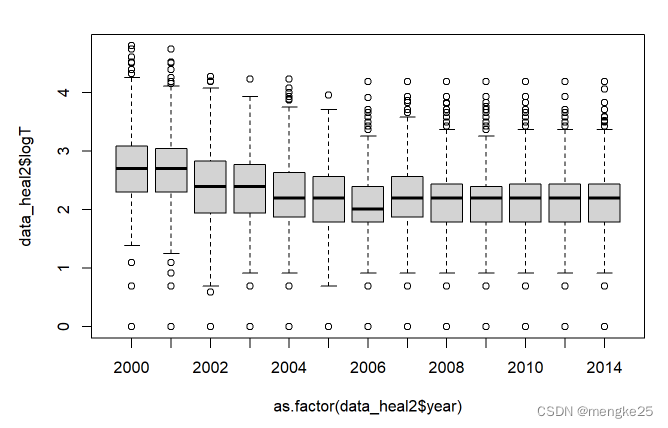
9.dataclear in hs8-year level
data0 <- data[data$capt == 0 ,]
data0_P <- data.frame(aggregate(data0$lnP,list(data0$Year,data0$hs8),mean))
data0_Q <- data.frame(aggregate(data0$lnQ,list(data0$Year,data0$hs8),mean))
data0_V <- data.frame(aggregate(data0$lnV,list(data0$Year,data0$hs8),mean))
names(data0_P) <-c("year","hs","meanP")
names(data0_Q) <-c("year","hs","meanQ")
names(data0_V) <-c("year","hs","meanV")
data0_ <- merge(data0_P,data0_Q,by.x = c("year","hs"),by.y = c("year","hs"))
data0_ <- merge(data0_,data0_V,by.x = c("year","hs"),by.y = c("year","hs"))
data1 <- subset(data,data$capt == 1 )
data1_P <- data.frame(aggregate(data1$lnP,list(data1$Year,data1$hs8),mean))
data1_Q <- data.frame(aggregate(data1$lnQ,list(data1$Year,data1$hs8),mean))
data1_V <- data.frame(aggregate(data1$lnV,list(data1$Year,data1$hs8),mean))
names(data1_P) <-c("year","hs","meanP")
names(data1_Q) <-c("year","hs","meanQ")
names(data1_V) <-c("year","hs","meanV")
data1_ <- merge(data1_P,data1_Q,by.x = c("year","hs"),by.y = c("year","hs"))
data1_ <- merge(data1_,data1_V,by.x = c("year","hs"),by.y = c("year","hs"))
data2 <- subset(data,data$capt == 2 )
data2_P <- data.frame(aggregate(data2$lnP,list(data2$Year,data2$hs8),mean))
data2_Q <- data.frame(aggregate(data2$lnQ,list(data2$Year,data2$hs8),mean))
data2_V <- data.frame(aggregate(data2$lnV,list(data2$Year,data2$hs8),mean))
names(data2_P) <-c("year","hs","meanP")
names(data2_Q) <-c("year","hs","meanQ")
names(data2_V) <-c("year","hs","meanV")
data2_ <- merge(data2_P,data2_Q,by.x = c("year","hs"),by.y = c("year","hs"))
data2_ <- merge(data2_,data2_V,by.x = c("year","hs"),by.y = c("year","hs"))
data0_$BEC <- 0
data1_$BEC <- 1
data2_$BEC <- 2
data_ <- rbind(data0_,data1_,data2_)
rm(data0_P,data0_Q,data0_V,data1_P,data1_Q,data1_V,data2_P,data2_Q,data2_V,data0,data1,data2)
data_$lnmeanQ <- log(data_$meanQ)
data_$lnmeanP <- log(data_$meanP)
data_$lnmeanV <- log(data_$meanV)
10.scatter
ggplot(data_, aes(lnmeanP, lnmeanV, color = BEC))+
geom_point(size = 2.0, shape = 16)
data_cha <- data.frame(subset((data_),data_$BEC != 0))
ggplot(data_cha, aes(lnmeanP, lnmeanV, color = BEC))+
geom_point(size = 2.0, shape = 16)
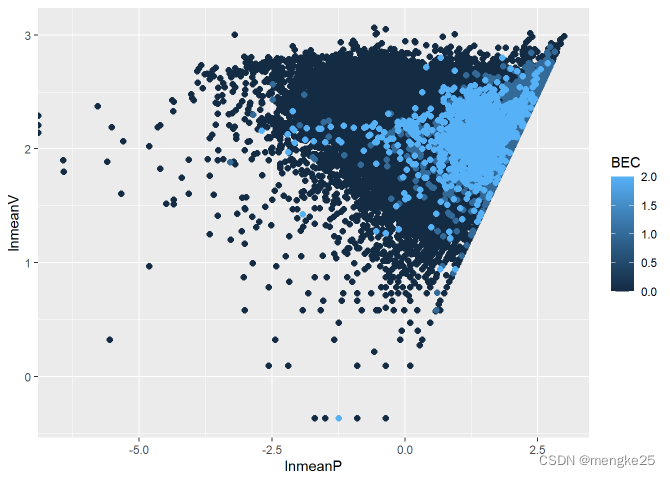
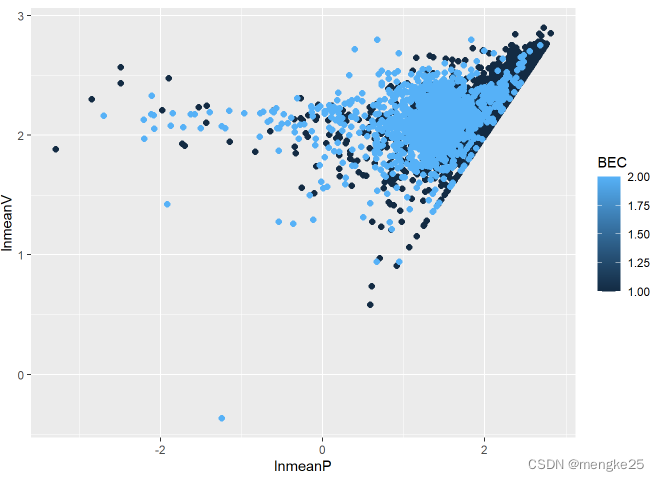
11.density
11.1density-type1
p <- ggplot(data = data_, mapping = aes(
x = lnmeanV, y = year))
p + geom_jitter(width = 0, height = 0.3, alpha = 0.5)
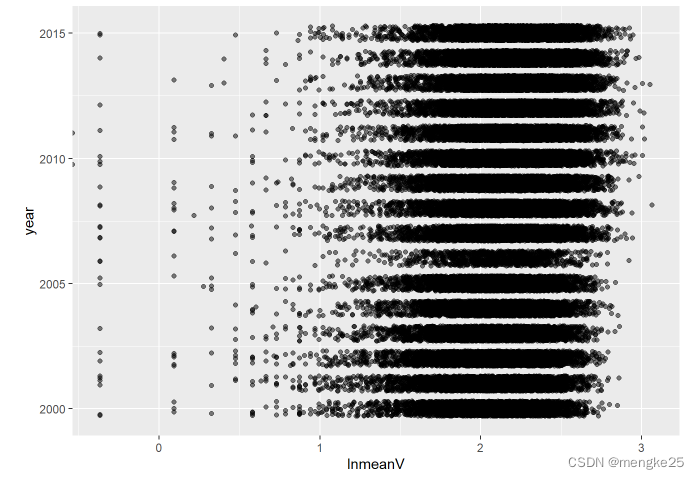
11.2density-type2
11.2.1 no-shade
p <- ggplot(data = data_, mapping = aes(
x = lnmeanV, color = year))
p + geom_line(stat = "density")
data_$year_str <- as.character(data_$year)
p <- ggplot(data = data_, mapping = aes(
x = lnmeanV, color = year_str))
p + geom_line(stat = "density")
11.2.2 shade
p <- ggplot(data = data_, mapping = aes(
x = lnmeanV, color = year_str, fill = year_str))
p + geom_density(alpha = 0.4)
12.summarize the data
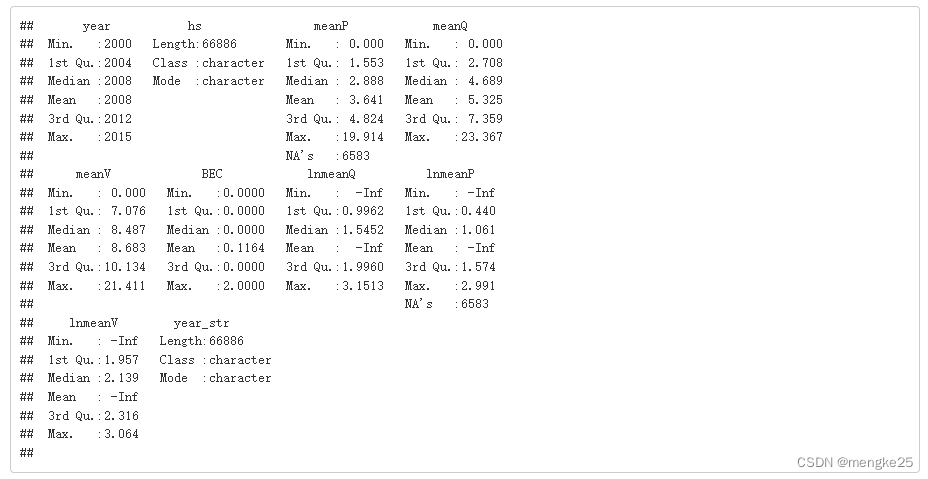
summary(data_,maxsum = 20, quantile.type = 7)
13.regress
n <- 10
x <- rnorm(n,1)
y <- rnorm(n,1)
fit_try <- lm(y ~ x)
summary(fit_try)
data_$ifNA <- complete.cases(data_)
data_reg <- as.data.frame(subset(data_,data_$ifNA == TRUE))
# fit <- lm(lnmeanV ~ lnmeanP, data =data_)
# summary(fit)
fit2 <- lm(lnmeanV ~ lnmeanP, data =data_[data_$BEC==1, ])
summary(fit2)
fit3 <- lm(lnmeanV ~ lnmeanP, data =data_[data_$BEC !=0, ])
summary(fit3)
fit4 <- estimatr::lm_robust(lnmeanV ~ lnmeanP , data = data_[data_$BEC !=0, ], se_type= "stata")
summary(fit4)
fit5 <- estimatr::lm_robust(lnmeanV ~ lnmeanP , data = data_[data_$BEC !=0, ], clusters = hs)
summary(fit5)





14.reg R versus stata
# 简单回归
# reg lwage i.numdep
# reg lwage c.educ#c.exper
# reg lwage c.educ##c.exper
# reg lwage c.exper##i.numdep
#
# lm(lwage ~ as.factor(numdep), data= wage1)
# lm(lwage ~ educ:exper, data =wage1)
# lm(lwage ~ educ*exper, data =wage1)
# lm(wage ~ exper*as.factor(numdep),data = wage1)
fit6 <- estimatr::lm_robust(lnmeanV ~ as.factor(year) , data = data_[data_$BEC !=0, ], se_type= "stata")
summary(fit6)
ggcoefstats(fit6,exclude.intercept = TRUE,stats.labels = FALSE)
# coefplot
# p <- ggplot(data = ddsmall_summ, mapping = aes(
# x = cut,
# y = mean_carat ))
# p + geom_point(size = 1.2) +
# geom_pointrange(mapping = aes(
# ymin = mean_carat - sd_carat,
# ymax = mean_carat + sd_carat)) +
# labs(x = "Cut",
# y = "Carat")
#### 面板回归
# xtset id year
# xtdescribe
# xtsum
# xtreg mrdrte unem, fe
#
# plm::is.pbalanced(murder$id,murder$year)
# modfe <- plm::plm(mrdrte ~ unem,index = c("id", "year"),model ="within", data = murder)
# summary(modfe)
15.dataclear for bar & line
Data0_P <- data.frame(aggregate(data0_$meanP,list(data0_$hs),mean))
Data0_Q <- data.frame(aggregate(data0_$meanQ,list(data0_$hs),mean))
Data0_V <- data.frame(aggregate(data0_$meanV,list(data0_$hs),mean))
Data1_P <- data.frame(aggregate(data1_$meanP,list(data1_$hs),mean))
Data1_Q <- data.frame(aggregate(data1_$meanQ,list(data1_$hs),mean))
Data1_V <- data.frame(aggregate(data1_$meanV,list(data1_$hs),mean))
Data2_P <- data.frame(aggregate(data2_$meanP,list(data2_$hs),mean))
Data2_Q <- data.frame(aggregate(data2_$meanQ,list(data2_$hs),mean))
Data2_V <- data.frame(aggregate(data2_$meanV,list(data2_$hs),mean))
names(Data0_P) <- c("hs","meanP")
names(Data0_Q) <- c("hs","meanQ")
names(Data0_V) <- c("hs","meanV")
names(Data1_P) <- c("hs","meanP")
names(Data1_Q) <- c("hs","meanQ")
names(Data1_V) <- c("hs","meanV")
names(Data2_P) <- c("hs","meanP")
names(Data2_Q) <- c("hs","meanQ")
names(Data2_V) <- c("hs","meanV")
Data0_ <- merge(Data0_P,Data0_Q,by.x = c("hs") ,by.y = c("hs"))
Data0_ <- merge(Data0_,Data0_V,by.x = c("hs") ,by.y = c("hs"))
Data1_ <- merge(Data1_P,Data1_Q,by.x = c("hs") ,by.y = c("hs"))
Data1_ <- merge(Data1_,Data1_V,by.x = c("hs") ,by.y = c("hs"))
Data2_ <- merge(Data2_P,Data2_Q,by.x = c("hs") ,by.y = c("hs"))
Data2_ <- merge(Data2_,Data2_V,by.x = c("hs") ,by.y = c("hs"))
rm(Data0_P,Data0_Q,Data0_V,Data1_P,Data1_Q,Data1_V,Data2_P,Data2_Q,Data2_V)
Data0_P <- data.frame(aggregate(data0_$meanP,list(data0_$year),mean))
Data0_Q <- data.frame(aggregate(data0_$meanQ,list(data0_$year),mean))
Data0_V <- data.frame(aggregate(data0_$meanV,list(data0_$year),mean))
Data1_P <- data.frame(aggregate(data1_$meanP,list(data1_$year),mean))
Data1_Q <- data.frame(aggregate(data1_$meanQ,list(data1_$year),mean))
Data1_V <- data.frame(aggregate(data1_$meanV,list(data1_$year),mean))
Data2_P <- data.frame(aggregate(data2_$meanP,list(data2_$year),mean))
Data2_Q <- data.frame(aggregate(data2_$meanQ,list(data2_$year),mean))
Data2_V <- data.frame(aggregate(data2_$meanV,list(data2_$year),mean))
names(Data0_P) <- c("year","meanP")
names(Data0_Q) <- c("year","meanQ")
names(Data0_V) <- c("year","meanV")
names(Data1_P) <- c("year","meanP")
names(Data1_Q) <- c("year","meanQ")
names(Data1_V) <- c("year","meanV")
names(Data2_P) <- c("year","meanP")
names(Data2_Q) <- c("year","meanQ")
names(Data2_V) <- c("year","meanV")
DatA0_ <- merge(Data0_P,Data0_Q,by.x = c("year") ,by.y = c("year"))
DatA0_ <- merge(DatA0_,Data0_V,by.x = c("year") ,by.y = c("year"))
DatA1_ <- merge(Data1_P,Data1_Q,by.x = c("year") ,by.y = c("year"))
DatA1_ <- merge(DatA1_,Data1_V,by.x = c("year") ,by.y = c("year"))
DatA2_ <- merge(Data2_P,Data2_Q,by.x = c("year") ,by.y = c("year"))
DatA2_ <- merge(DatA2_,Data2_V,by.x = c("year") ,by.y = c("year"))
rm(Data0_P,Data0_Q,Data0_V,Data1_P,Data1_Q,Data1_V,Data2_P,Data2_Q,Data2_V)
16.bar
ggplot(DatA2_, aes(x=year, y=meanQ))+
geom_bar(position=position_dodge(), stat="identity") +
scale_y_continuous(breaks=0:20*4) +
scale_x_continuous(breaks=0:10*0.5)+
theme_bw()+
theme(panel.grid.major=element_line(colour=NA),
panel.background = element_rect(fill = "transparent",colour = NA),
plot.background = element_rect(fill = "transparent",colour = NA),
panel.grid.minor = element_blank(),legend.justification=c(0.85,0.1),legend.position=c(0.85,0.1))
DatA0_$BEC <- 0
DatA1_$BEC <- 1
DatA2_$BEC <- 2
DatA <- rbind(DatA0_,DatA1_,DatA2_)
ggplot(DatA,aes(x=year,y=meanQ,fill=BEC))+
geom_bar(stat='identity', position='stack') +
theme_bw() +
labs(x = 'Specie',y = 'Value', title = 'This is barplot') +
theme(axis.title =element_text(size = 16),axis.text =element_text(size = 14, color = 'black'),plot.title =element_text(hjust = 0.5, size = 20))
ggplot(DatA,aes(x=year,y=meanQ,fill=BEC))+
geom_bar(stat='identity', position='dodge') +
theme_bw() +
labs(x = 'Specie',y = 'Value', title = 'This is barplot') +
theme(axis.title =element_text(size = 16),axis.text =element_text(size = 14, color = 'black'),plot.title =element_text(hjust = 0.5, size = 20))
DatA$BEC_str <- as.character(DatA$BEC)
ggplot(DatA,aes(x=year,y=meanQ,fill=BEC_str))+geom_bar(position="dodge",stat="identity")+
xlab("年份") + ylab("数量") + labs(fill="类型")
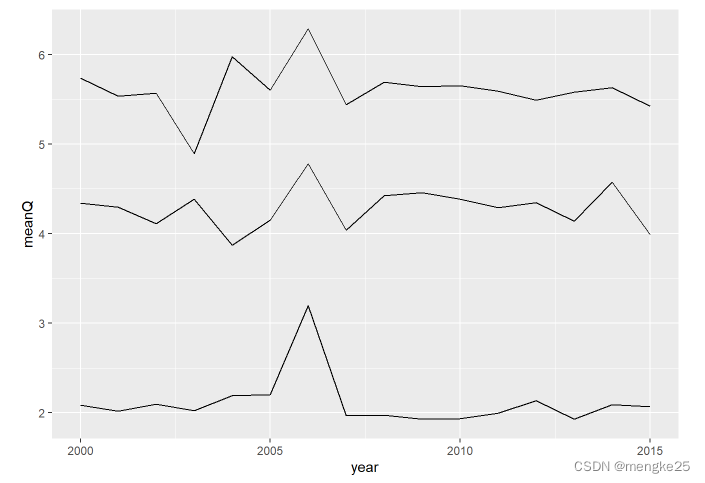
17.line
ggplot(DatA, aes(year, meanQ,group=BEC))+
geom_path(lineend = "butt", linejoin = "round", stat = "identity",linemitre = 10) +
geom_line(stat = "identity",position = "identity" )